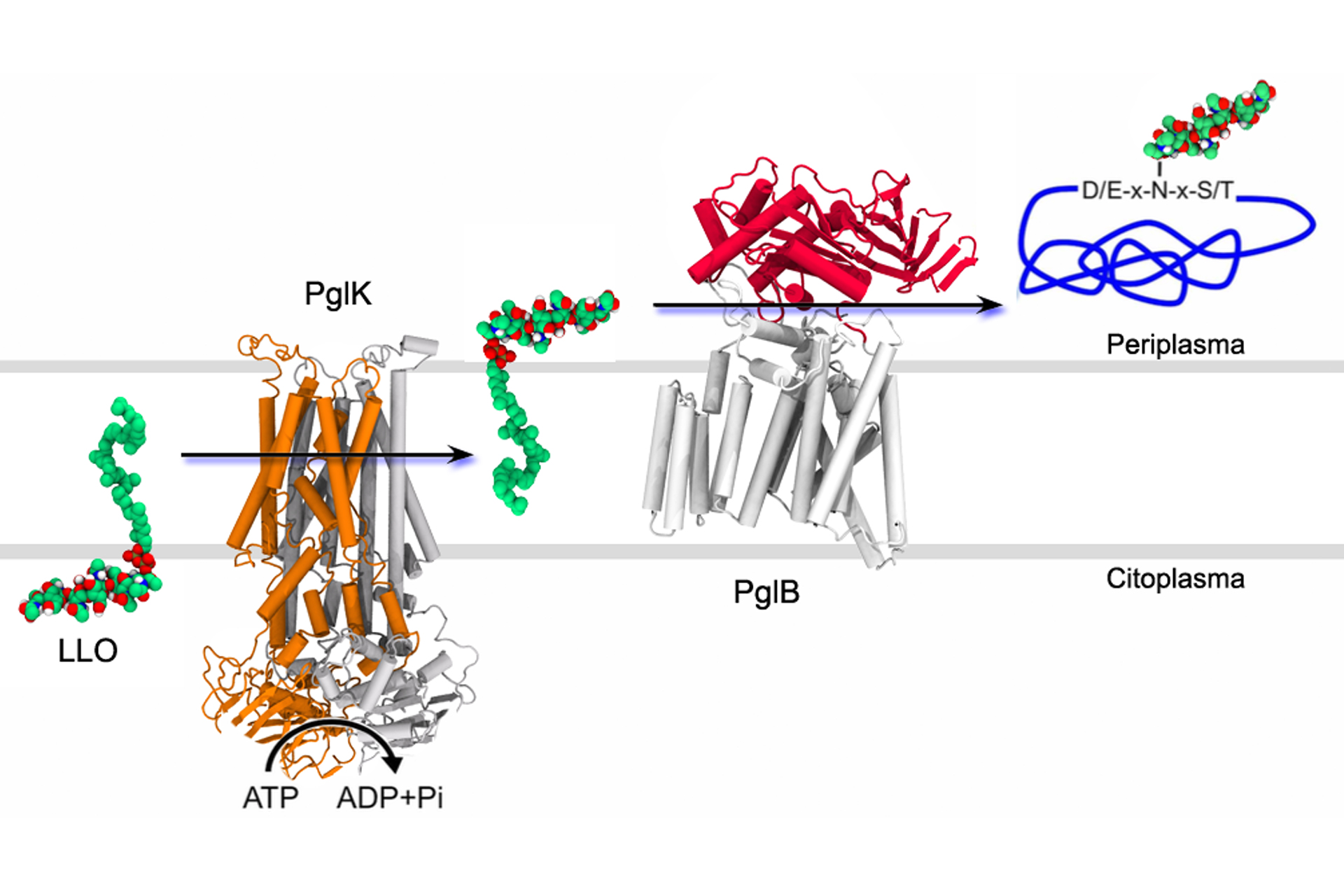
About Me
My name is Pablo Ricardo Arantes, and I am currently a research scientist at an R&D company that develops new pharmaceutical products and generic ones for use in humans, working within three technological platforms: Drug Delivery System, Biotechnology, and Organic Synthesis.
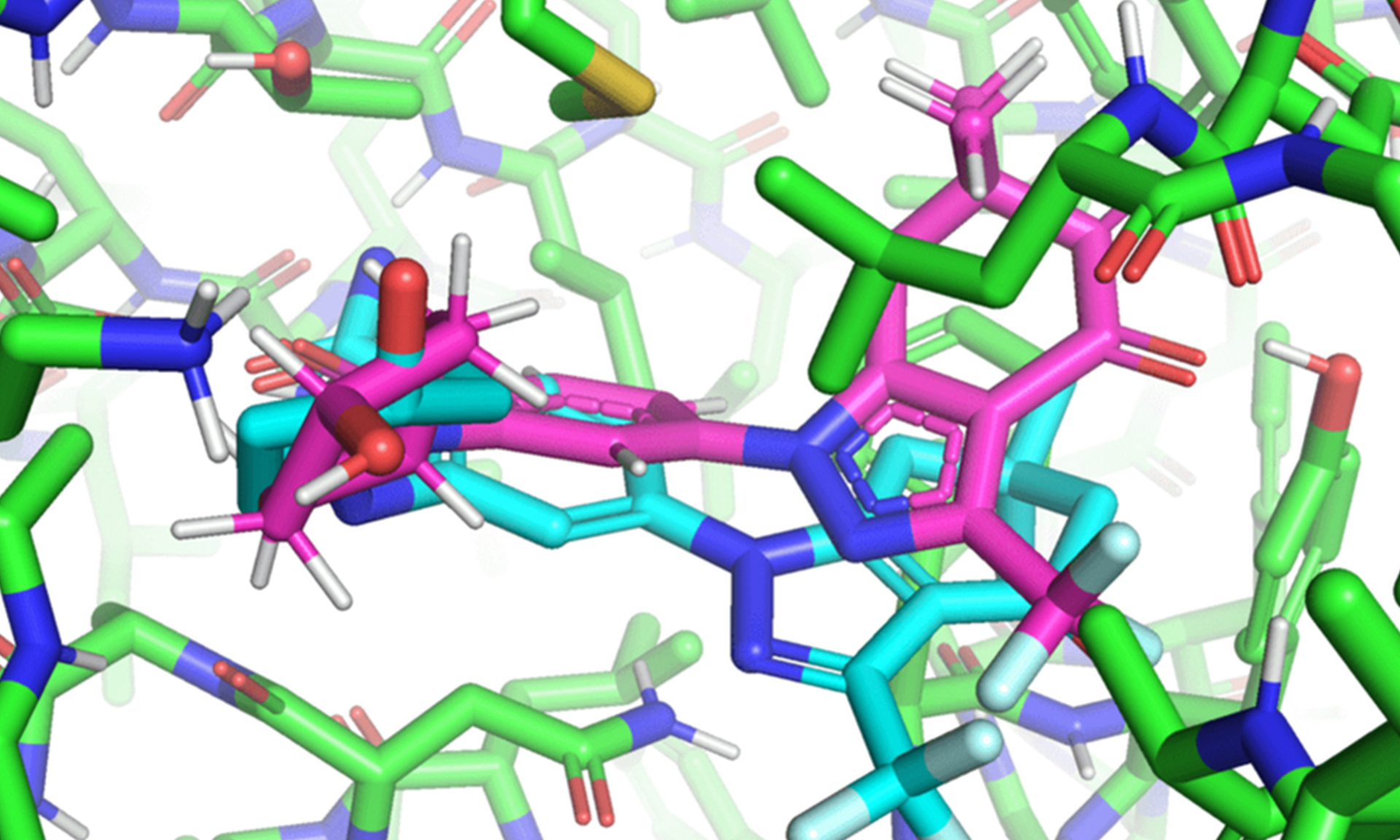
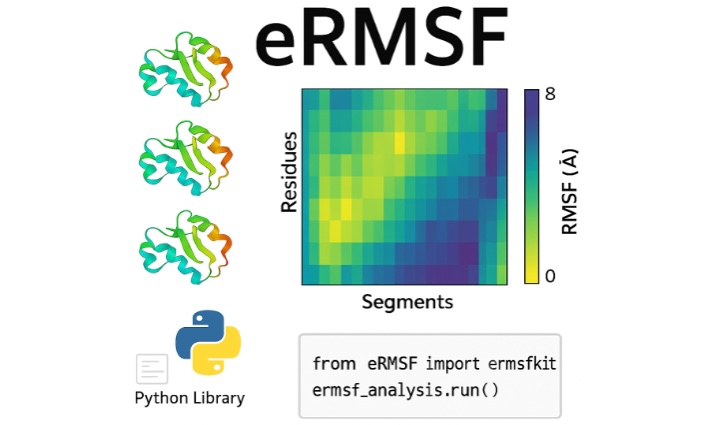
Previously, I worked at the University of California, Riverside, in the field of biophysics (Palermo Lab). I hold extensive expertise in computational biophysics and knowledge in state‐of‐the‐art computational methods that have been proven instrumental in delving deep into biological function.
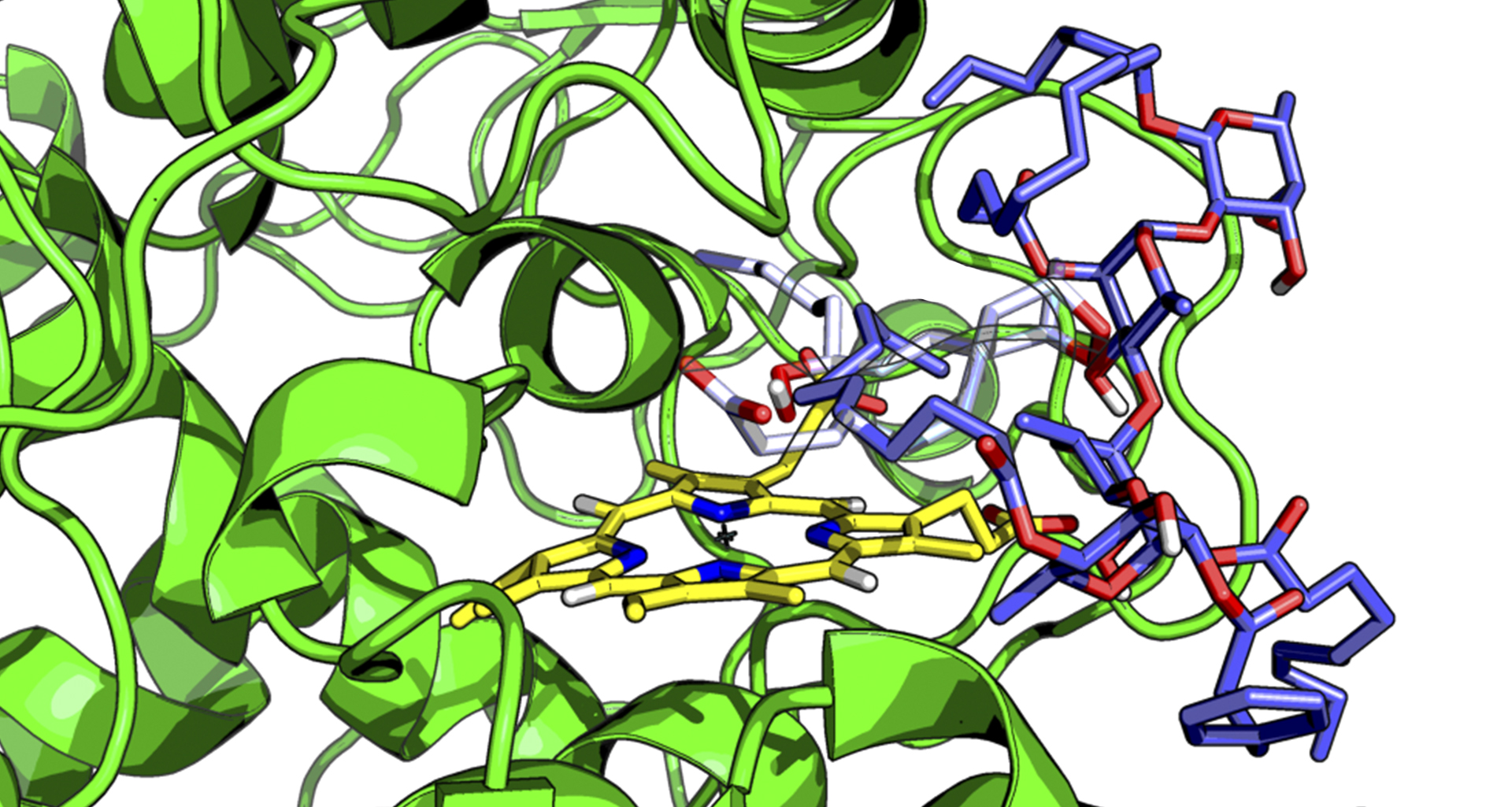
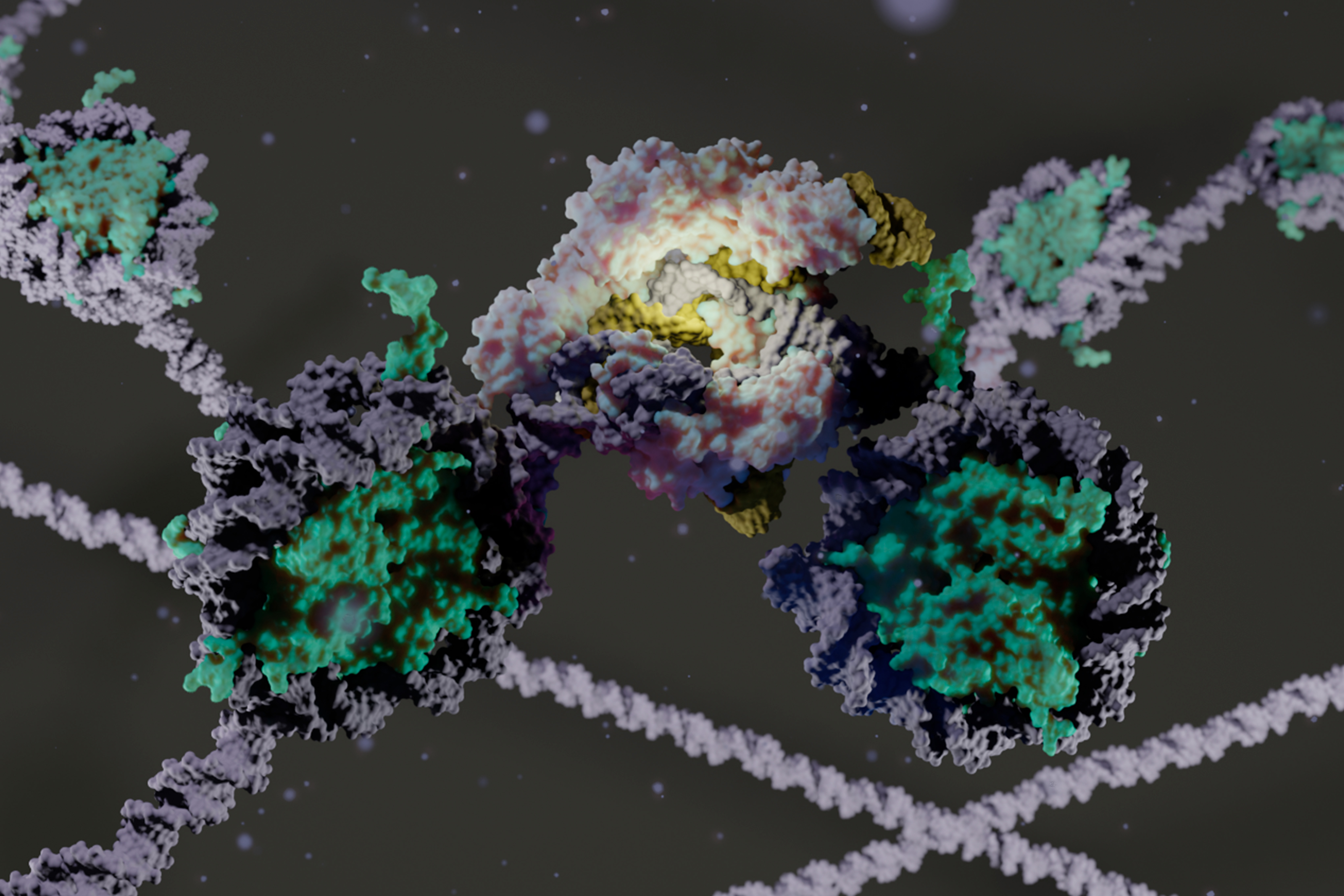
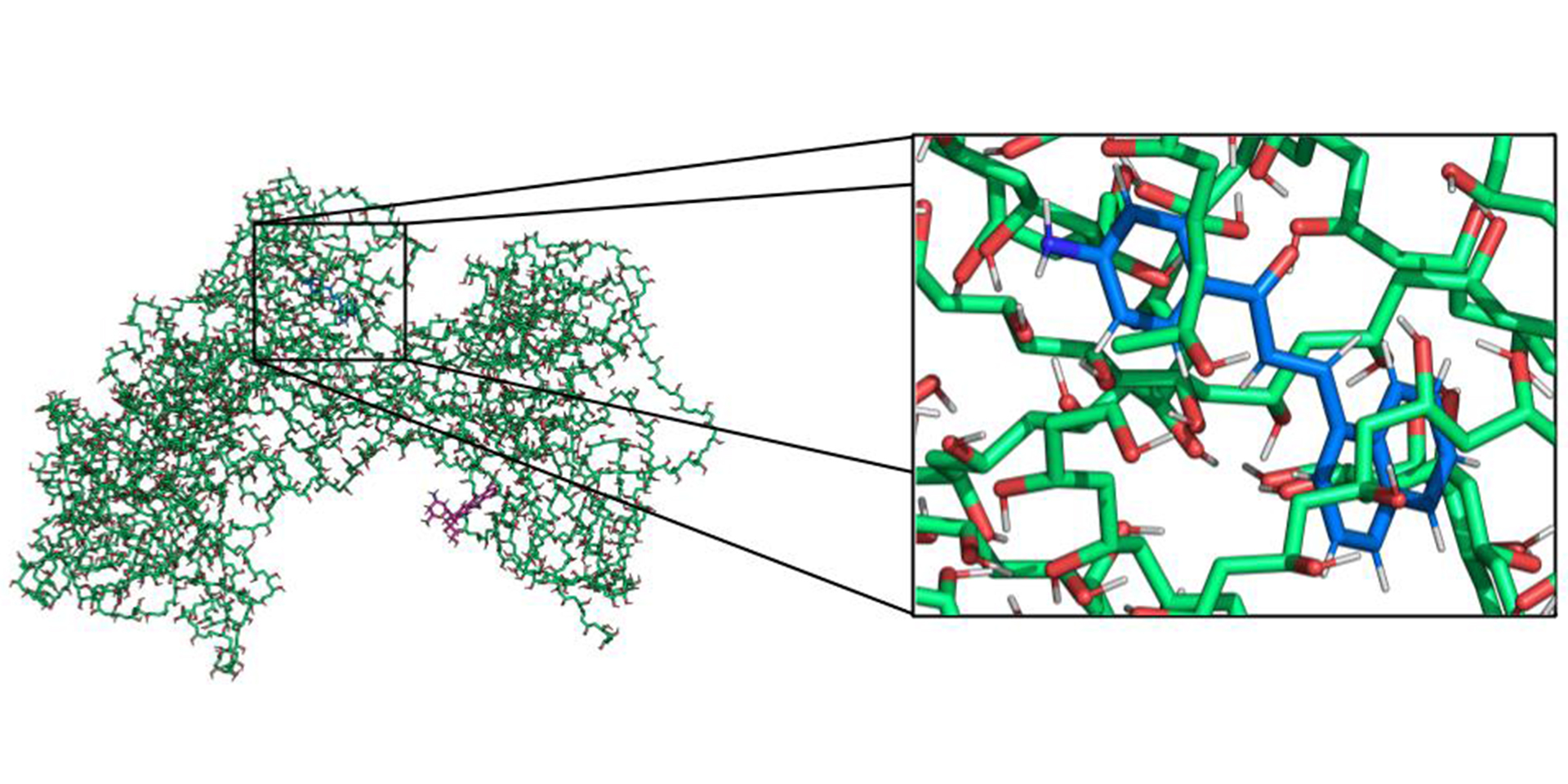
My research at the University of California focused on molecular simulations of emerging genome editing technologies based on the CRISPR‐Cas9 system, which received the Nobel Prize in Chemistry 2020.
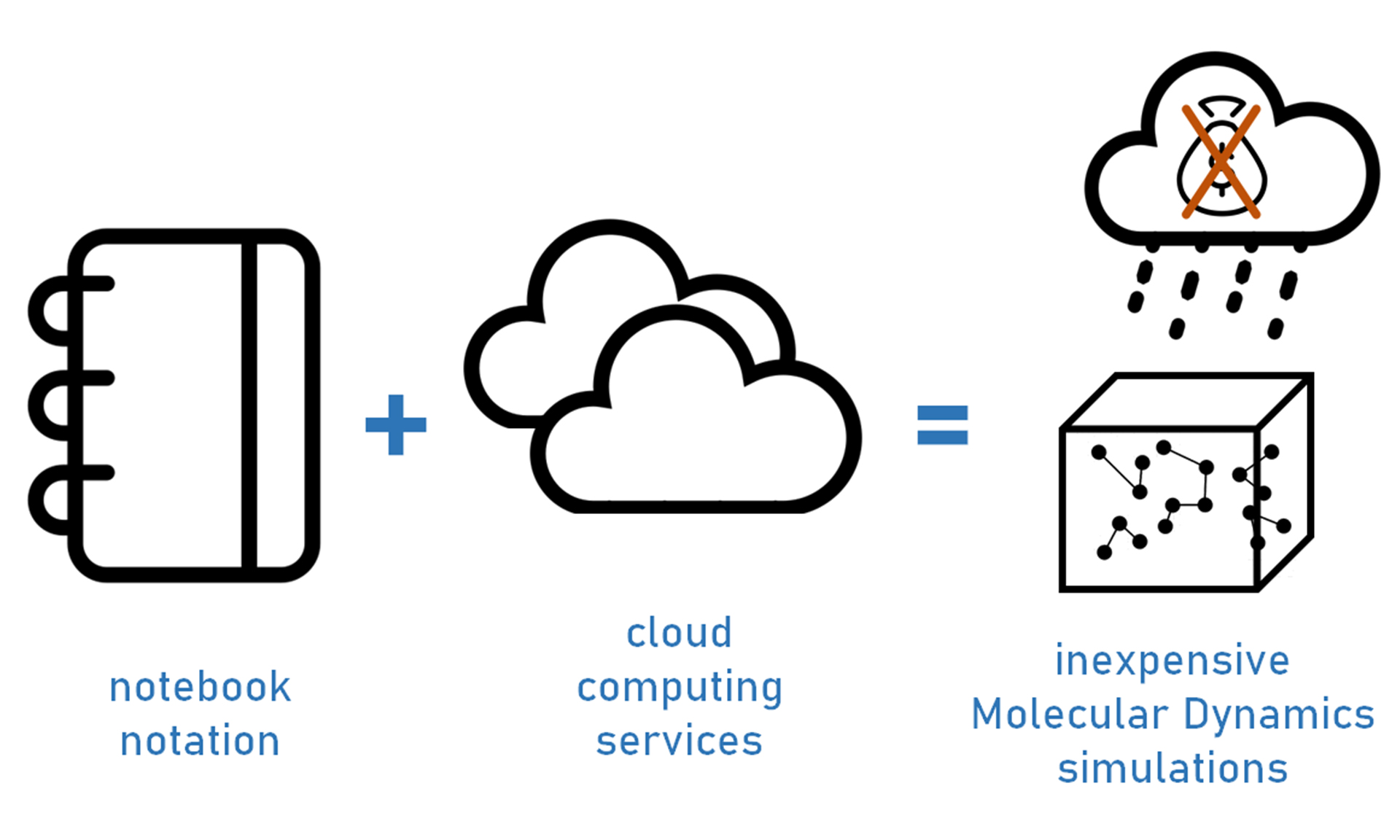
In 2021, I worked on the first user‐friendly front‐end for running molecular dynamics (MD) simulations using the OpenMM toolkit on the Google Colab framework (Making it rain). An affordable (cost‐free) strategy for performing molecular dynamics simulations using cloud computing resources. It involves shareable, ready‐to‐use, customizable Jupyter notebooks that guide the end‐user in running their calculations using Google Colab services.
My current collaboration network includes biochemists, bioinformaticians, and structural biologists from Brazil, Uruguay, England, Spain, Australia, and the USA.